
De novo design and experimental characterization of ultrashort self-associating peptides
J. Smadbeck, K.H. Chan, G.A. Khoury, B. Xue, R.C. Robinson, C.A.E. Hauser, and C.A. Floudas
PLOS Computational Biology 10 [7] (2014) e1003718
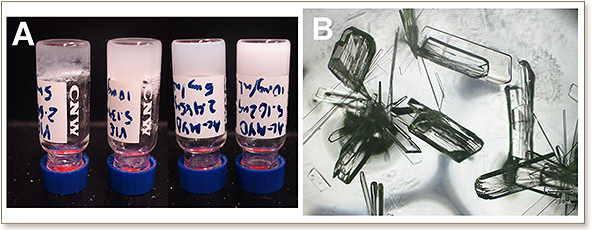
Self-association is a common phenomenon in biology and one that can have positive and negative impacts, from the construction of the architectural cytoskeleton of cells to the formation of fibrils in amyloid diseases. Understanding the nature and mechanisms of self-association is important for modulating these systems and in creating biologically-inspired materials. Here, we present a two-stage de novo peptide design framework that can generate novel self-associating peptide systems. The first stage uses a simulated multimeric template structure as input into the optimization-based Sequence Selection to generate low potential energy sequences. The second stage is a computational validation procedure that calculates Fold Specificity and/or Approximate Association Affinity (K*(association)) based on metrics that we have devised for multimeric systems. This framework was applied to the design of self-associating tripeptides using the known self-associating tripeptide, Ac-IVD, as a structural template. Six computationally predicted tripeptides (Ac-LVE, Ac-YYD, Ac-LLE, Ac-YLD, Ac-MYD, Ac-VIE) were chosen for experimental validation in order to illustrate the self-association outcomes predicted by the three metrics. Self-association and electron microscopy studies revealed that Ac-LLE formed bead-like microstructures, Ac-LVE and Ac-YYD formed fibrillar aggregates, Ac-VIE and Ac-MYD formed hydrogels, and Ac-YLD crystallized under ambient conditions. An X-ray crystallographic study was carried out on a single crystal of Ac-YLD, which revealed that each molecule adopts a beta-strand conformation that stack together to form parallel beta-sheets. As an additional validation of the approach, the hydrogel-forming sequences of Ac-MYD and Ac-VIE were shuffled. The shuffled sequences were computationally predicted to have lower K*(association) values and were experimentally verified to not form hydrogels. This illustrates the robustness of the framework in predicting self-associating tripeptides. We expect that this enhanced multimeric de novo peptide design framework will find future application in creating novel self-associating peptides based on unnatural amino acids, and inhibitor peptides of detrimental self-aggregating biological proteins.
DOI: 10.1371/journal.pcbi.1003718

"KAUST shall be a beacon for peace, hope and reconciliation, and shall serve the people of the Kingdom and the world."
King Abdullah bin Abdulaziz Al Saud, 1924 – 2015
Thuwal 23955-6900, Kingdom of Saudi Arabia
Al-Haytham Building (Bldg. 2)
© King Abdullah University of Science and Technology. All rights reserved